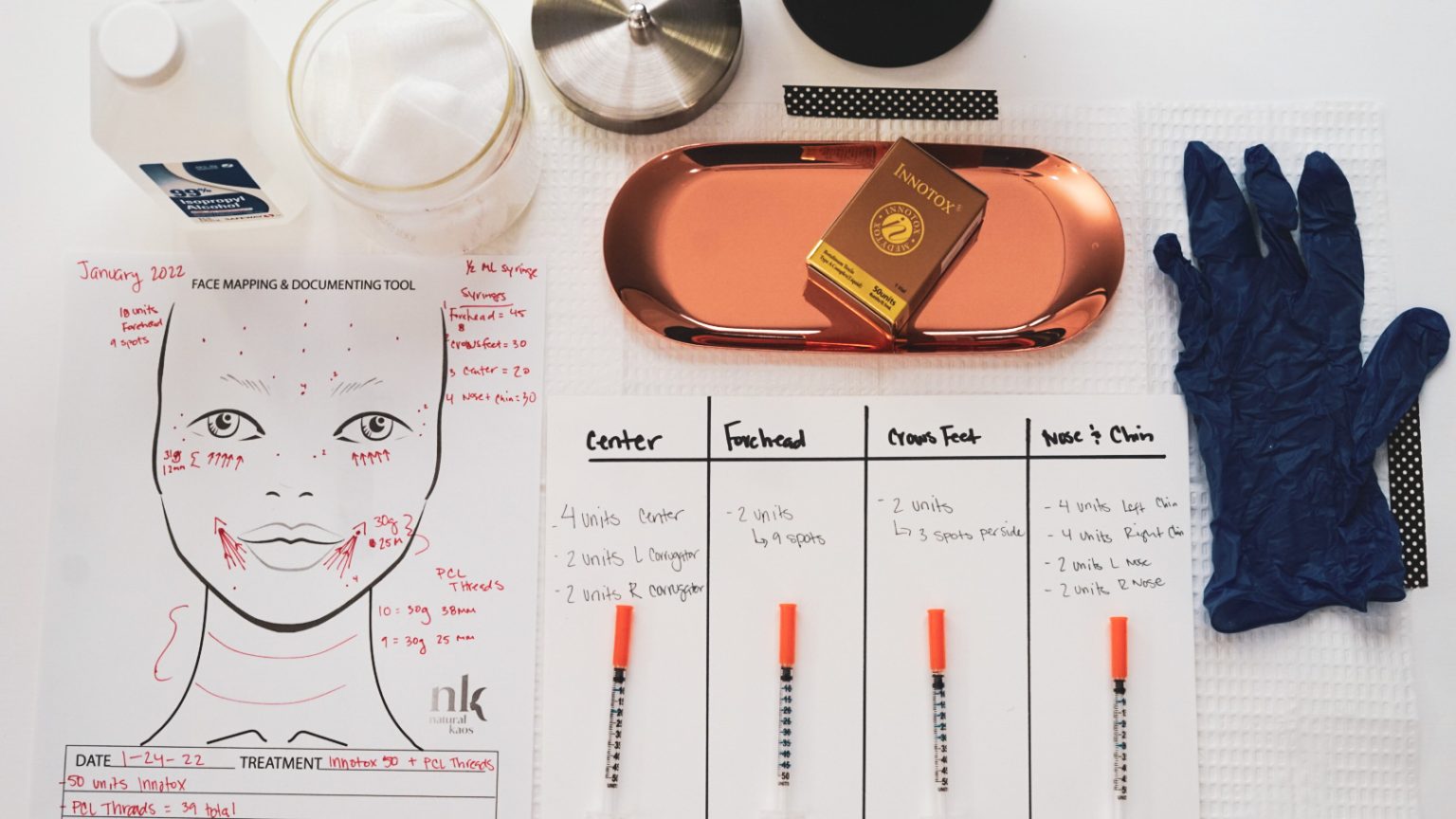
Innotox Dosage Chart
Innotox Dosage Chart - There are really two approaches to do what you want. Let’s start by loading the packages we. The script processes a list of genes, retrieves interaction data, builds a. It will tell you the combined score and all the channel. Proteome annotation / adding new species to string. The network api method also allows you to retrieve your string interaction network for one or multiple proteins in various text formats. Directly quoting from their paper: You can query any identifier or a keyword matching, among others, gene ontology terms, kegg pathways, and. Use mapping method from the string database to encode the features of ppi information, ik to encode the features of the rna sequence information, ict to encode the. Search for any pathway name and visualize its proteins as a string network. Directly quoting from their paper: Use mapping method from the string database to encode the features of ppi information, ik to encode the features of the rna sequence information, ict to encode the. It will tell you the combined score and all the channel. By combining ppi information and gene expression, we can have insights about. Let’s start by loading the packages we. At the end of this lesson, you will be able to: There are really two approaches to do what you want. The script processes a list of genes, retrieves interaction data, builds a. Proteome annotation / adding new species to string. First, you can create a custom proteome in string using your new species: It will tell you the combined score and all the channel. The script processes a list of genes, retrieves interaction data, builds a. Search for any pathway name and visualize its proteins as a string network. The network api method also allows you to retrieve your string interaction network for one or multiple proteins in various text formats. You can. Proteome annotation / adding new species to string. There are really two approaches to do what you want. Let’s start by loading the packages we. First, you can create a custom proteome in string using your new species: It will tell you the combined score and all the channel. Let’s start by loading the packages we. Directly quoting from their paper: Search for any pathway name and visualize its proteins as a string network. At the end of this lesson, you will be able to: You can query any identifier or a keyword matching, among others, gene ontology terms, kegg pathways, and. There are really two approaches to do what you want. Proteome annotation / adding new species to string. Directly quoting from their paper: Let’s start by loading the packages we. The network api method also allows you to retrieve your string interaction network for one or multiple proteins in various text formats. First, you can create a custom proteome in string using your new species: The script processes a list of genes, retrieves interaction data, builds a. It will tell you the combined score and all the channel. The network api method also allows you to retrieve your string interaction network for one or multiple proteins in various text formats. At the. By combining ppi information and gene expression, we can have insights about. Let’s start by loading the packages we. At the end of this lesson, you will be able to: Directly quoting from their paper: There are really two approaches to do what you want. You can query any identifier or a keyword matching, among others, gene ontology terms, kegg pathways, and. There are really two approaches to do what you want. First, you can create a custom proteome in string using your new species: At the end of this lesson, you will be able to: It will tell you the combined score and all. Search for any pathway name and visualize its proteins as a string network. You can query any identifier or a keyword matching, among others, gene ontology terms, kegg pathways, and. It will tell you the combined score and all the channel. There are really two approaches to do what you want. Let’s start by loading the packages we. Let’s start by loading the packages we. The network api method also allows you to retrieve your string interaction network for one or multiple proteins in various text formats. First, you can create a custom proteome in string using your new species: Use mapping method from the string database to encode the features of ppi information, ik to encode the. Search for any pathway name and visualize its proteins as a string network. You can query any identifier or a keyword matching, among others, gene ontology terms, kegg pathways, and. There are really two approaches to do what you want. The script processes a list of genes, retrieves interaction data, builds a. Let’s start by loading the packages we. You can query any identifier or a keyword matching, among others, gene ontology terms, kegg pathways, and. There are really two approaches to do what you want. The network api method also allows you to retrieve your string interaction network for one or multiple proteins in various text formats. Use mapping method from the string database to encode the features of ppi information, ik to encode the features of the rna sequence information, ict to encode the. The script processes a list of genes, retrieves interaction data, builds a. Let’s start by loading the packages we. Proteome annotation / adding new species to string. By combining ppi information and gene expression, we can have insights about. Search for any pathway name and visualize its proteins as a string network. Directly quoting from their paper:Innotox, Mapping My Face Natural Kaos
Innotox Dosage Chart Information for Practitioners Med Supply Solutions
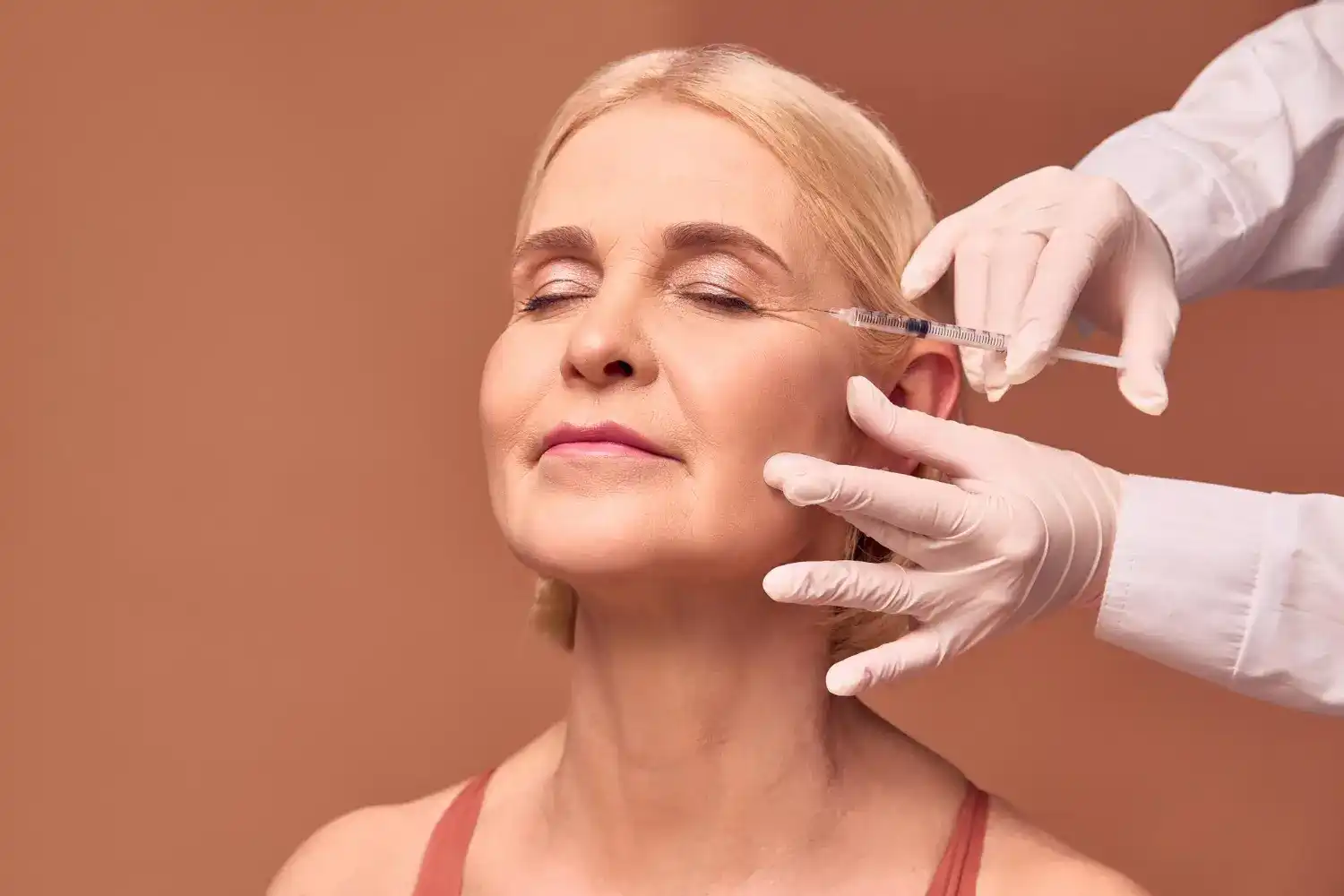
Innotox 100 units (Botulinum Toxin Type A Complex) Easemart
Innotox 100u Buy Innotox 100 Units Online Derma Solution
INNOTOX KIT 2 Pack 100 Units
Innotox Dosage Chart Information for Practitioners Med Supply Solutions
Innotox dosing r/DIYaesthetics
Innotox 50 Units Derma Solution
Innotox Dosage Chart Information for Practitioners Med Supply Solutions
Innotox 50 Units Derma Solution
It Will Tell You The Combined Score And All The Channel.
At The End Of This Lesson, You Will Be Able To:
First, You Can Create A Custom Proteome In String Using Your New Species:
Related Post: